-
1. Introduction
2. CPC 2 Online Input
3. CPC 2 Results Output
3.2 CPC 2 results: raw data
3.3 CPC 2 results details
3.4 CPC 2 Evidence Output
1. IntroductionBack to top
Coding Potential Calculator distinguishes protein-coding from non-coding RNAs based on the sequence features of the input transcripts. Last version of CPC1 is widely used by worldwide researchers. For better serve for scientific community, we update CPC1 to CPC2. It can discriminate the coding and non-coding transcripts faster and more accurately. We provide an online version of CPC2 here.
2. CPC2 Online InputBack to top
CPC2 accepts RNA transcript sequences as input. Both fasta format and GTF/GFF/BED format are supported.
2.1 Submit requirement
Fasta format:
Size requirement: Less than 100000 lines in input box and no line limitation in batch model. Maximum allowable upload file size is 50 Mb
Name requirement: Sequence names beginning with ‘>’symbol are required. ID characters after blank character will be discarded in the results.
Sequence requirement: Only characters in DNA and RNA sequences are allowed.
GTF/GFF/BED format:
Both BED6, BED12, GTF and GFF format are supported.
Less than 50000 lines. Maximum allowable upload file size is 50 Mb
GTF/GFF/BED file of following genome assembly are allowed:
Human (hg38), Human (hg19), Chimpanzee (panTro4), Mouse (mm10), Rat (rn6), Zebrafish (danRer7), Xenopus (xendTro3), Fruitfly (dm6)
NOTE: Input BED format will slow down the speed. Ensure your file is NO larger than 50 Mb and 10,000 lines.
2.2 How to submit?
There are 2 ways to submit lncRNA sequences:
Paste lncRNA sequences into the big input box at the home page.
Upload fasta file by the batch operation.
2.3 Additional Options
Also check the reverse complement strand:
Coding potential of both positive strand RNA and negative strand RNA will be assessed.
NOTE: The reverse complement strand check is NOT a part of the CPC 2 algorithm. We make the service available here to provide additional information for users. And switching on this option might slow the speed a little.
3. CPC 2 Results OutputBack to top
The results will be stored on our server for one month, you can retrive the result via our "Batch Page" during the time.
3.1 CPC 2 results: html view
CPC 2 results html view gives an overview of coding status of the input sequences. Each row corresponds to one input sequence. The columns show the sequence ID, the coding/noncoding classification label, the coding probability (the "distance" to the SVM classification hyper-plane in the features space), scores of three features (putative peptide length, Fickett TESTCODE score, putative isoelectric point),rhe ORF integrity and the "Details" link (as described later).
3.2 CPC 2 results: raw data
Raw data of CPC 2 output results could be downloaded by clicking on "Download the result" button.
Raw data contains 9 columns are separated by tab, and each line stands for the result of an input sequence. For example:

The columns show the sequence ID, putative peptide length, Fickett score, isoelectric point, ORF start position, the integrity of the orf, coding probability and the coding/noncoding classification label.
3.3 CPC 2 results details
Results details consist of a summary paragraph, a graphical view of detail information in feature distribution of your sequence and more information of additional evidences for coding potential.
Detail information in feature distribution
We provide a graphic view of details in three features calculated by CPC 2 and its distribution in known coding/noncoding RNAs.
The background color area of the three images depict a total distribution of features in coding/noncoding RNAs. Blue area refers to features calculated from known noncoding transcripts in Ensembl database. Yellow area refers to features calculated from mRNAs in RefSeq database with SwissProt annotation.
Move mouse along the area edge to see a distribution frequency between the specific interval. Click on the button “Noncoding/Coding” above the graph to show or hide the distribution area of Noncoding/Coding RNAs as screenshot shows.
The black column shows the feature value of your input sequence and its position in the background distribution interval. And whether it points the coding distribution or noncoding distribution relies on the classification answer given by CPC 2.

You can download the graph by clicking on the right button.

Additional evidences for coding
To further help users to assess coding ability of input RNA sequences, we provided the BLAST searching against SwissProt, RNAdb, lncRNAdb as a supplementary module for CPC 2 web server.
Parameters for SwissProt:
blastx -evalue 1e-10 -ungapped -num_threads 2 -outfmt 6 -comp_based_stats F -query seq.fasta -db swissprot -out output_swissprot
Parameters for RNAdb and lncRNAdb:
blastn -num_threads 2 -outfmt 6 -query seq.fasta -db rnadb.fa -out output_rnadb
3.4 CPC 2 Evidence Output
Users can view CPC 2 evidence results in its raw data format and in html format.
Html view
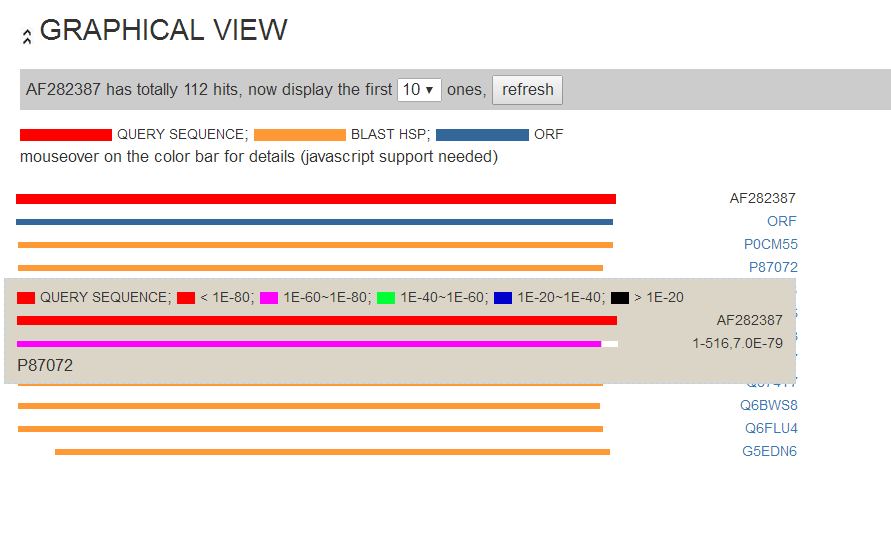
We provide a graphical view of predicted peptide and BLAST results summary. This page shows the position of putative ORF, and BLAST hsp.
Mousing over the ORF and blast hsp color bar, users can view details of predicted ORF and BLAST hits such as BLAST e-value of the hits.
Raw data format
Blast results in tabular format:
| 1. | qseqid | query (e.g., gene) sequence id |
| 2. | sseqid | subject (e.g., reference genome) sequence id |
| 3. | pident | percentage of identical matches |
| 4. | length | alignment length |
| 5. | mismatch | number of mismatches |
| 6. | gapopen | number of gap openings |
| 7. | qstart | start of alignment in query |
| 8. | qend | end of alignment in query |
| 9. | sstart | start of alignment in subject |
| 10. | send | end of alignment in subject |
| 11. | evalue | expect value |
| 12. | bitscore | bit score |
| 13. | databse | the databse of this query |
4. FAQBack to top
How do I retrieve results?
When user gives input sequences to the CPC2 web server for calculating, the web server will assign an unique Task ID to this request. After finishing the calculating, users can go to the Batch Page and use the Task ID to retrieve their results from CPC2 server.
What's new in CPC2?
In the updated CPC2, we employ a novel discriminative model based on sequence intrinsic features, which effectively addresses the two issues: the CPC2 runs two orders of magnitude faster than CPC1, and also be the fastest tool among other popular ones ; Meanwhile, the CPC2 shows superior accuracy than CPC1. The last but not the least, the updated model in CPC2, as in the CPC1, is species-neutral, making it feasible/accessible for the ever-growing non-model-organism transcriptomes.